
Open-Source Internship opportunity by OpenGenus for programmers. Apply now.
Reading time: 30 minutes
The Bron–Kerbosch algorithm is an enumeration algorithm for finding maximal cliques in an undirected graph. That is, it lists all subsets of vertices with the two properties that each pair of vertices in one of the listed subsets is connected by an edge, and no listed subset can have any additional vertices added to it while preserving its complete connectivity.
The Bron–Kerbosch algorithm was designed by Dutch scientists Coenraad Bron and Joep Kerbosch, who published its description in 1973.
The Bron–Kerbosch algorithm is not an output-sensitive algorithm. Unlike some other algorithms for the clique problem, it does not run in polynomial time per maximal clique generated. However, it is efficient in a worst-case sense: by a result of Moon & Moser (1965), any n-vertex graph has at most 3n⁄3 maximal cliques, and the worst-case running time of the Bron–Kerbosch algorithm (with a pivot strategy that minimizes the number of recursive calls made at each step) is O(3n⁄3).
Algorithm
The algorithm maintains three disjoint sets of nodesR, P, X. Set R stands for the currently growing clique; set P stands for prospective nodes which are connected to all nodes in R and using which R can be expanded; set X contains nodes already processed i.e. nodes which were previously in P and hence all maximal cliques containing them have already been reported. An important invariant is that all nodes which are connected to every node of R are either in P or X. Purpose of X is to avoid reporting the same maximal clique multiple times, through the update P = P − {ui}. To avoid reporting cliques which are not maximal, the algorithm checks whether set X is empty, if X is non-empty, the nodes in X may be added to R, but this would yield previously found maximal cliques.
Without pivoting
The basic form of the Bron–Kerbosch algorithm is a recursive backtracking algorithm that searches for all maximal cliques in a given graph G. More generally, given three disjoint sets of vertices R, P, and X, it finds the maximal cliques that include all of the vertices in R, some of the vertices in P, and none of the vertices in X. In each call to the algorithm, P and X are disjoint sets whose union consists of those vertices that form cliques when added to R. In other words, P ∪ X is the set of vertices which are joined to every element of R. When P and X are both empty there are no further elements that can be added to R, so R is a maximal clique and the algorithm outputs R.
BronKerbosch1(R, P, X):
if P and X are both empty:
report R as a maximal clique
for each vertex v in P:
BronKerbosch1(R ⋃ {v}, P ⋂ N(v), X ⋂ N(v))
P := P \ {v}
X := X ⋃ {v}
With pivoting
The basic form of the algorithm, described above, is inefficient in the case of graphs with many non-maximal cliques, it makes a recursive call for every clique, maximal or not. To save time and allow the algorithm to backtrack more quickly in branches of the search that contain no maximal cliques, Bron and Kerbosch introduced a variant of the algorithm involving a "pivot vertex" u, chosen from P (or more generally, as later investigators realized, from P ⋃ X). Any maximal clique must include either u or one of its non-neighbors, for otherwise the clique could be augmented by adding u to it. Therefore, only u and its non-neighbors need to be tested as the choices for the vertex v that is added to R in each recursive call to the algorithm.
BronKerbosch2(R,P,X):
if P and X are both empty:
report R as a maximal clique
choose a pivot vertex u in P ⋃ X
for each vertex v in P \ N(u):
BronKerbosch2(R ⋃ {v}, P ⋂ N(v), X ⋂ N(v))
P := P \ {v}
X := X ⋃ {v}
With vertex ordering
An alternative method for improving the basic form of the Bron–Kerbosch algorithm involves forgoing pivoting at the outermost level of recursion, and instead choosing the ordering of the recursive calls carefully in order to minimize the sizes of the sets P of candidate vertices within each recursive call.
The degeneracy of a graph G is the smallest number d such that every subgraph of G has a vertex with degree d or less. Every graph has a degeneracy ordering, an ordering of the vertices such that each vertex has d or fewer neighbors that come later in the ordering; a degeneracy ordering may be found in linear time by repeatedly selecting the vertex of minimum degree among the remaining vertices. If the order of the vertices v that the Bron–Kerbosch algorithm loops through is a degeneracy ordering, then the set P of candidate vertices in each call (the neighbors of v that are later in the ordering) will be guaranteed to have size at most d. The set X of excluded vertices will consist of all earlier neighbors of v, and may be much larger than d. In recursive calls to the algorithm below the topmost level of the recursion, the pivoting version can still be used.
BronKerbosch3(G):
P = V(G)
R = X = empty
for each vertex v in a degeneracy ordering of G:
BronKerbosch2({v}, P ⋂ N(v), X ⋂ N(v))
P := P \ {v}
X := X ⋃ {v}
The exact running time of the algorithm depends on implementation details. The number of recursive calls to the procedure. By improving the algorithm, we optimize the number of recursive calls in "without pivoting method" with "with pivoting method", thus it improves the runtime complexity of the algorithm.
For further improvement of running time of the algorithm we introduce vertex ordering in the outermost level of the recursion, which optimizes the overall size of set P,thus using pivot strategy and vertex ordering gives the worst case running time of O(3n⁄3). This is optimal as a function of n, since there exist up to 3n⁄3 maximal cliques in an n-vertex graph.
Implementation
Code in Python3
class Node(object):
def __init__(self, name):
self.name = name
self.neighbors = []
def __repr__(self):
return self.name
A = Node('A')
B = Node('B')
C = Node('C')
D = Node('D')
E = Node('E')
F = Node('F')
A.neighbors = [B, C, E]
B.neighbors = [A, C, D, F]
C.neighbors = [A, B, D, F]
D.neighbors = [C, B, E, F]
E.neighbors = [A, D]
F.neighbors = [B, C, D]
all_nodes = [A, B, C, D, E, F]
def find_cliques(potential_clique=[], remaining_nodes=[], skip_nodes=[], depth=0):
if len(remaining_nodes) == 0 and len(skip_nodes) == 0:
print('This is a clique:', potential_clique)
return 1
found_cliques = 0
for node in remaining_nodes:
# Try adding the node to the current potential_clique to see if we can make it work.
new_potential_clique = potential_clique + [node]
new_remaining_nodes = [n for n in remaining_nodes if n in node.neighbors]
new_skip_list = [n for n in skip_nodes if n in node.neighbors]
found_cliques += find_cliques(new_potential_clique, new_remaining_nodes, new_skip_list, depth + 1)
# We're done considering this node. If there was a way to form a clique with it, we
# already discovered its maximal clique in the recursive call above. So, go ahead
# and remove it from the list of remaining nodes and add it to the skip list.
remaining_nodes.remove(node)
skip_nodes.append(node)
return found_cliques
total_cliques = find_cliques(remaining_nodes=all_nodes)
print('Total cliques found:', total_cliques)
Output
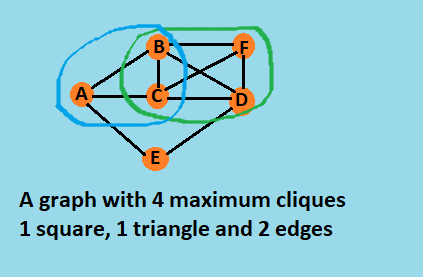
This is a clique: [A, B, C]
This is a clique: [A, E]
This is a clique: [C, B, D, F]
This is a clique: [E, D]
Total cliques found: 4
Complexity
Time Complexity
- Bron-Kerbosch algorithm takes O(3n⁄3) time in the worst case.
Space Complexity
- Bron-Kerbosch algorithm takes O(n2) auxiliary space in the worst case.
Related articles
Greedy approach to find a single maximal clique in O(V^2) time complexityAlgorithm to find cliques of a given size k 【O(n^k) time complexity】
